This article was first published in Molecular Plant Breeding (2012, 10(4): 457-461) in Chinese, and here was authorized to translate and publish the paper in English under the terms of Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Wild rice,
Oryza longistaminata, originated in Africa, belongs to AA genome the genus Oryza, which is a wild perennial species with the characteristics of cross-pollination, self-incompatibility and underground stems. The
Oryza Longisatminata has obvious performance of drought tolerance (
Liu et al., 2004) and as well as bacterial blight resistance. One of the well-known blight resistant gene,
Xa21, had been cloned through the positional cloning approach (
Khush et al., 1990;
Song et al., 1995).
Genomic library is an important material platform for the study of genomics and functional genomics. Currently, large fragment genomic library has been widely used for new gene mining and gene function analysis in animals, plants and microbes.
Fosmid is a vector system constructed by Kim and his colleagues who introduced the pBAC into PUCcos, the length of the inserted fragment closes with the cosmid (
Kim et al., 1992). Fosmid library plays an important role in constructing physical map (
Birren et al., 1996;
Fitz-Gibbon et al., 1997;
Magrini et al., 2004), map-based cloning (d
e Tomaso and Weissman, 2003) and alignment analysis (
Larkin et al., 2003;
Newman et al., 2005), etc.
The sequencing of the whole genome sequence of the Nipponbare, Fosmid library also be used to fill the gaps in the genome sequence (
Ammiraju et al., 2005). Although the inserted fragment in Fosmid library is more small than that of bacterial artificial chromosome library (BAC), Fosmid library possesses some advantages in good stability, no bias, building cycle short and simple operation, which would be a good platform for quickly building a library, cloning and sequencing new gene as well as other applications.
In this study, we attempt to build a high-quality Fosmid library of wild African rice whole genome (Oryza longistaminata) in order to provide an excellent research tool for exploring and utilizing the favorable genes in the Oryza Longisatminata as well as cloning genes.
1 Results and analysis
1.1 Genomic DNA preparation
Genomic DNA was extracted by using the modified CTAB method, and total 100 μg DNA with the concentration of 5 μg/μL was obtained in this research. The size of DNA fragment is mainly more than 30 kb in length measured by pulsed-field gel electrophoresis (
Figure 1), DNA fragments framed in the figure were the DNA in the range from 36 to 48 kb in length we needed were recovered, which the DNA concentration and total amount within this range sufficient met the requirements of the follow-up experiments on DNA quality, it didn’t need to do the DNA broken treatment.
|

Figure 1 DNA concentration detected by the pulsed-field gel electrophoresis (PFGE)
|
1.2 The insertion fragment and stability detected by restriction endonuclease digestion
Thirty one clones were randomly picked up to be detected their plasmid by restriction endonuclease NotI digenstion. The digested products were detected by Pulsed-field gel electrophoresis to calculated their average insert size of 41.7 kb in length (Figure 2), and also appeared a 7.5 kb vector fragment. There was no empty vector happened in the detection, and the size of the insertion meet the requirements for constructing Fosmid library.
|
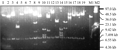
Figure 2 Fosmid clones digested with restriction enzyme NotI
|
BGI, the sequencing company, assessed the insertion sizes of the 13 351 clones and calculated about 40 kb of the average insertion size in this library; 12 644 clones of them accounting for 94.7% had the insertion fragment with the size between 30~50 kb in length (
Figure 3).
|
.png)
Figure 3 Distribution of insertion fragments in size
|
Clones with continuous regeneration, the clones of the initiative generation and the 100
th generation were digested by restriction endonuclease
EcoRI and
HindIII, validating that no digesting bands lost and or banding rearrangement happened (
Figure 4), which indicated that the constructed Fosmid library was very stable.
|
.png)
Figure 4 Stability assays of Fosmid clones
|
1.3 Titer and total number of clones calculated
Titer was calculated following up the formula, that is, Titer (CFU/mL) = (number of colonies×dilution times ×103 μL/mL)/amount of diluting unamplified library (μL). Titer of this library was 5.6×104 CFU/ mL. Total 6 mL mixed packages were obtained in this research, expecting to get total 336 000 clones, in which we picked monoclonal 111 360 placed on 290 plates with 384-well each plate, and stored at -80℃ refrigerator. Therefore, it is possible to estimate the remaining non picked mixed packages contained about 220 000 clones that were used for end sequencing.
1.4 Analysis of library coverage
In this research we picked up and stored 111 360 clones with the average insertion size of 40 kb, assumption the size of the rice genome being about 430 Mb (
Arumuganathan and Earle, 1991) , the library covered 10.4 times of whole rice genome. Based on Clarke-Carbon’s formula, the probability of a gene optionally screened from the library was 99.99%, which needed 99 007 clones. So the library we built had 99.99% probability of any gene to be screened.
1.5 Super pool screening
In this study, total six microsatellite markers tightly linked with underground stems, of which Rhz10, Rhz12 and Rhz13 on chromosome 3, and Rhz82, Rhz85 and Rhz86 on chromosome 4, were employed to screen the super pools.
Figure 5 presented that the number of super pools screened by using PCR with six markers. As we can be seen that numbers of the super pools screened with six markers were from 9 to 27 in range, in other words, at least positive clones 9~27 should be screened that indicated the library constructed in this research should have excellent practice.
|
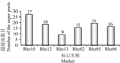
Figure 5 Figure 5 The super pools screened by using SSR markers
|
1.6 End Sequencing
Except for clones the library for permanently preservation, the remaining mixed packages of the about 220 000 clones were carried out in paired-end sequencing. After removal of the sequences of vectors as well as repetitive sequences, 124 700 effective terminal sequences were obtained, the effective read length each sequence was greater than 44 bp. Terminal sequencing made an important contribution to assembly and splice of the re- sequencing wild rice whole genome of wild rice. Since introduction of the sequence of fosmid sequencing, it was not only to reduce the number of scaffold, but also extended the length of the scaffold (unpublished data).
2 Discussion
DNA preparation is the first step to construct a library. The size requirement of DNA fragments in Fosmid library is around 40 kb, we abandoned the complicated approaches for preparing high molecular weight DNA instead of adopting universal CTAB extraction method that the extracted DNA would be able to meet the require- ments of DNA fragment size for constructing the library without doing the DNA breaking treatment.
Preparation of high molecular weight DNA has been one of the important steps as well as the basis for constructing a high quality library. However, obtaining a high molecular weight DNA was often time-consuming and labor-cost because of the cumbersome preparing steps and rigorous operating requirement.
Fosmid library do not need to digest with restriction enzymes, and the carrying capacity was about 40 kb. If the high molecular weight DNA is too long, then that needs to do shearing treatment so as to achieve a 40 kb length DNA fragment. The numbers of searing treatment also need to be tested, the less treatment might generate long DNA fragments, while the more treatment will lead to generate short DNA fragments. We employed the popular CTAB method that obviated the tedious steps and saved time, it was more convenient as well.
The insert size of the fragments and the extent of coverage of the genome are the key for measuring whether or not the quality of the Fosmid library is good or bad. The Fosmid library constructed in this research were fully consistent with the construction requirements of Fosmid library, such as the size of the inserted fragment and the stability. Therefore, the library constructed in this research possessed high quality that fully meets the requirements of the gene mining and sequencing.
So far, it might be the first report on the cons- truction of Fosmid library in wild rice, Oryza longistaminata; it was the first application by using the combination with the Fosmid end sequencing and the next-generation sequencing technology to the extension and splicing of the genome as well. This wild rice whole genome fosmid library will also be provided free to the researchers who are doing associated work. At the same time, the applications combined with next generation sequen- cing technology led to a new direction in the application of the fosmid library as well as broadened the insight of the fosmid library development.
3 Materials and methods
3.1 Research Materials
The wild rice (Oryza longistaminata) used in this research collected and preserved in Yunnan Academy of Agricultural Sciences. The fresh leaves were sampled for whole-genome DNA extraction.
3.2 Major chemicals
Library construction kit (Cat.No.CCFOS059) was developed by the Epicentre company. Other reagents, such as medium, were imported reagents or domestic analytical grade chemicals.
3.3 Research Methods
3.3.1 genomic DNA preparation
Directly using the modified CTAB method (
Murray and Thompson, 1980) to extract DNA. Taking 2 g of fresh leaves, then adding liquid nitrogen and fully grinding. The ground powder was then quickly transferred to the centrifuge tube containing 2×CTAB solution, wherein 0.1 g powder by adding 0.5 mL 2×CTAB solution, placing on water bath at 55℃ for 10 min. Taking the supernatant after centrifuged at 12 000 r/min for 5 min. Adding chloroform and isoamyl alcohol (24:1), shocking on oscillator 1 min. Again, taking the supernatant after centrifuged at 12 000 r/min for 5 min. Adding 100 μL sodium acetate with 7.5 mol/L at pH 7.0 and 1 mL ice-ethanol, mixing and then standing 10 min at -20℃. Discarding the supernatant after centri- fuged at 12 000 r/min for 5 min. Adding 1 mL 70% ethanol to be washed 2 times, discarding the supernatant, and then dried. Finally, DNA was dissolved by adding 100 μL 0.5×TE solution. Taking a little DNA sample for measuring DNA fragment size by using the pulsed field gel electrophoresis under the conditions as 1% low melting point agarose gel and 0.5×TBE electrophoresis buffer, 6 V/cm electronic voltage and kept the temperature at 14℃, while the pulse conversion time was set as 5 s~15 s, electrophoresis was run about 16 hours.
3.3.2 Terminal modification of DNA fragment
Blunt-end and the 5'phosphorylation of DNA fragments were carried out by using terminal modification reagents provided in the kit. The terminal modification will enhance the efficiency and accuracy of connection with vector. 80 μL reaction system was designed in this experiment with the reaction at room temperature for 45 min and inactivation for 10 min at 70℃.
3.3.3 DNA fragment recovery
Terminal modified DNA was separated by pulsed-field gel electrophoresis, the size of the recovered fragments was around from 36 to 48 kb in length. Electrophoresis conditions were followed with 3.3.1.The ligation was reacted in volume of 10 μL at room temperature for 4 hours, and inactivated for 10 min at 70℃.
3.3.5 Packaging
The packaging protein (MaxPlax Lambda Packaging Extracts) was de-frozen on ice bath. Taking 25 μL packaging protein, and adding 10 μL ligation products, incubating at 30℃ for 90 min. Then adding 25 μL packaging and incubated for 90 min, 30℃. Finally, adding dilution solution, PDB, up to final volume 1 mL, and then adding 25 μL chloroform, slightly mixing and stored at 4℃ ready for use.
3.3.6 Determination of the titer of the packaging
The packaged phages were gradient diluted. Taking 10 μL of different gradient bacteriophage solution mixed with 100 μL of EPI300-T1R bacteria and incubated for 1 h, at 37℃. The incubated bacterium liquid was coated on the LB plate that contained chloramphenicol, and cultured overnight at 37℃. Titer was calculated based on the status of bacterium colony growth.
3.3.7 Clones picked and preserved
Single clone was picked and placed on the 384-well culture plate that contained a liquid LB medium with 12.5 μg/mL chloramphenicol and 20% glycerol, which were inoculated overnight at 37℃, and stored in refrigerator at -80℃.
3.3.8 library insert size and stability detection
Thirty one fosmid clones were picked up randomly, and the plasmid DNA was extracted by using the method of alkalinelysis, and then the plasmid DNA was digested by NotI.
The size of the inserted fragment was detected by the pulsed field gel electrophoresis.The inserted fragments of this library were consigned to be assessed by sequencing 13 351 clones to estimate the size of inserts in BGI.
Furthermore, five clones were randomly selected to be continuous cultured in the liquid LB medium containing chloramphenicol for 6 days, taking the bacteria at the first day (generation 0) and the sixth day (100 generations) used to extract plasmid DNA , respectively, then digested with restriction enzymes, EcoRI and HindIII, the stability of the inserted fragment detected by agarose gel electrophoresis.
3.3.9 Super pool building and initial screening
In this study, a 384-well plate bacteria were mixed into a sample as a super pool. In our study, six SSR markers tightly linked with underground stems gene were selected to screen the super pool by PCR approach (Kim et al, 2003).
3.3.10 Terminal sequencing
Except for the clones for permanent preservation, the remaining clones were mixed that was consigned to Beijing Genomics Institute (BGI) for terminal sequencing in order to assembly and splice of the re-sequencing wild rice whole genome of wild rice.
Author contributions
LLJ was the executor of the experimental design and experimental study; CJ and ZSL completed the data and results analysis; WL and HFY conceived this project and took responsible for the experimental design, data analysis, manuscript writing and modifying, who were the corresponding authors. All authors read and approved the final manuscript.
Acknowledgements
This research was funded by the National Natural Science Foundation (U0836605). The authors would like to thank Dr. Dai Luyuan in Yunnan Academy of Agricultural Sciences kindly provided the technical support in the course of the experiment and critical suggestion for the manuscript preparation.
References
Ammiraju J.S., Yu Y., Luo M., Kudrna D., Kim H., Goicoechea J.L., Katayose Y., Matsumoto T., Wu J., Sasaki T., and Wing R.A., 2005, Random sheared fosmid library as a new genomic tool to accelerate complete finishing of rice (
Oryza sativa spp. Nipponbare) genome sequence: sequencing of gap-specific fosmid clones uncovers new euchromatic portions of the genome, Theor. Appl. Genet., 111(8): 1596-1607 PMID: 16200416
Arumuganathan K., and Earle E.D., 1991, Nuclear DNA content of some important plant species, Plant Mol. Biol. Rep., 9(3): 208-218
Birren B.W., Tachi-iri Y., Kim U.J., Nguyen M., Shizuya H., Korenberg J.R., and Simon M.I., 1996, A human chromosome 22 fosmid resource: mapping and analysis of 96 clones, Genomics, 34(1): 97-106 PMID: 8661029
de Tomaso A.W., and Weissman I.L, 2003, Construction and characterization of large-insert genomic libraries (BAC and fosmid) from the Ascidian Botryllus schlosseri and initial physical mapping of a histocompatibility locus, Mar. Biotechnol. (NY), 5(2): 103-115
Fitz-Gibbon S.. Choi A.J., Miller J.H., Stetter K.O., Simon M.I., Swanson R., and Kim U.J., 1997, A fosmid-based genomic map and identification of 474 genes of the hyperthermophilic archaeon Pyrobaculum aerophilum, Extremophiles, 1(1): 36-51 PMID: 9680335
Khush G.S., Bacalangco E., and Ogawa T., 1990, A new gene for resistance to bacterial blight from O. longistaminata, Rice Genet. Newsl., 7: 121-122
Kim C.G., Fujiyama A., and Saitou N., 2003, Construction of a gorilla fosmid library and its PCR screening system, Genomics, 82(5): 571-574 PMID: 14559214
Kim U.J., Shizuya H., de Jong P.J., Birren B., and Simon M.I., 1992, Stable propagation of cosmid sized human DNA inserts in an F factor based vector, Nucl. Acids. Res., 20(5): 1083-1085 PMCID: PMC312094
Larkin D.M., Everts-van der Wind A., Rebeiz M., Schweitzer P.A., Bachman S., Green C., Wright C.L., Campos E.J., Benson L.D., Edwards J., Liu L., Osoegawa K., Womack J.E., de Jong P.J., and Lewin H.A., 2003, A cattle-human comparative map built with cattle BAC-ends and human genome sequence, Genome Res., 13(8): 1966-1972 PMCID: PMC403790
Liu L., Lafitte R., and Guan D., 2004, Wild Oryza species as potential sources of drought-adaptive traits, Euphytica, 138(2): 149-161
Magrini V., Warren W.C., Wallis J., Goldman W.E., Xu J., Mardis E.R., and McPherson J.D., 2004, Fosmid-based physical mapping of the Histoplasma capsulatum
genome, Genome Res., 14(8): 1603-1609
Murray M.G., and Thompson W.F., 1980, Rapid isolation of high molecular weight plant DNA, Nucl. Acids. Res., 8(19): 4321-4325 PMID: 7433111
Newman T.L., Tuzun E., Morrison V.A., Hayden K.E., Ventura M., McGrath S.D., Rocchi M., and Eichler E.E., 2005, A genome-wide survey of structural variation between human and chimpanzee, Genome. Res., 15(10): 1344-1356 PMCID: PMC1240076
Song W.Y., Wang G.L., Chen L.L., Kim H.S., Pi L.Y., Holsten T., Gardner J., Wang B., Zhai W.X., Zhu L.H., Fauquet C., and Ronald P., 1995, A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21, Science, 270(5243): 1804-1806
 Author
Author  Correspondence author
Correspondence author


.png)
.png)



